Weaker Immune Response to Viruses in Children with Mitochondrial Disorders
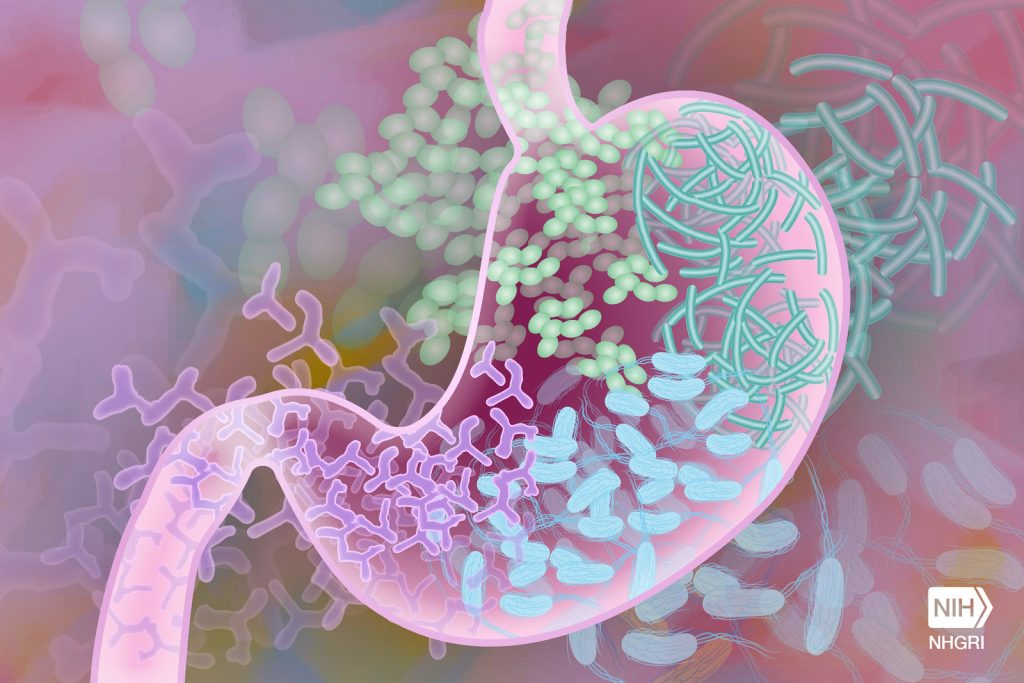
In a new study, National Institutes of Health (NIH) researchers found that altered B cell function in children with mitochondrial disorders led to a weaker and less diverse antibody response to viral infections. The study, published in Frontiers in Immunology, was led by researchers at the National Human Genome Research Institute (NHGRI), who analysed the gene activities of immune cells in children with mitochondrial disorders and found that B cells, which produce antibodies to fight viral infections, are less able to survive cellular stress.
“Our work is one of the first examples to study how B cells are affected in mitochondrial disease by looking at human patients,” said Eliza Gordon-Lipkin, MD, assistant research physician in NHGRI’s Metabolism, Infection and Immunity Section and co-first author of the paper.
Mitochondria are important components of nearly every cell in the body because they convert food and oxygen into energy. Genomic variants in more than 350 genes have been linked to mitochondrial disorders with varied symptoms depending on which cells are affected.
“For children with mitochondrial disorders, infections can be life threatening or they can worsen the progression of their disorder,” said Peter McGuire, MBBCh, NHGRI investigator, head of the Metabolism, Infection and Immunity Section and senior author of the study. “We wanted to understand how immune cells differ in these patients and how that influences their response to infections.”
Around 1 in 5000 people worldwide have a mitochondrial disorder. Examples of mitochondrial disorders are Leigh’s syndrome, which primarily affects the nervous system, and Kearns-Sayre syndrome, which primarily affects the eyes and heart.
While mitochondrial disorders are known to affect organs such as the heart, liver, and brain, less is known how they affect the immune system.
Using a genomic technique called single-cell RNA sequencing, which analyzes gene activity in different cell types, researchers studied immune cells found in blood. These cells include different types of white blood cells that help the body fight infections. During stressful conditions, these cells produce a microRNA called mir4485. MicroRNAs are small strings of RNA that help control when and where genes are turned on and off. mir4485 controls cellular pathways that help cells survive.
“We think that B cells in these patients undergo cellular stress when they turn into plasma cells and produce antibodies, and these B cells then try to survive by producing the microRNA to cope,” said Dr. McGuire. “But the B cells are too fragile due to their limited energy, so they are unable to survive the stressful conditions.”
Researchers used a technique called VirScan to look at all past viral infections, assess how well the immune system fought those infections and see the effects of B cells and plasma cells on antibody production. With a weaker antibody response, the immune systems in children with mitochondrial disorders are less able to recognize and neutralize invading viruses and clear infections.
Researchers aim to use the results of this study to guide future treatment of patients with mitochondrial disorders, noting that more translational studies are needed in this research area.
Source: National Institutes of Health