Nanoparticle and Antibiotic Polytherapy Defeats AMR Bacteria
Researchers from Monash University have discovered a potential new method to circumvent antibiotic resistance, by means of a nanoparticle and antibiotic polytherapy. This approach could also reduce antibiotic intake.
The World Health Organisation (WHO) has declared antimicrobial resistance (AMR) to be among the top 10 global public health threats. A recent report found that in 2019, 1.27 million deaths were directly attributable to AMR infections – more than deaths from either HIV or TB.
AMR occurs when pathogens evolve to no longer respond to medicines, consequently infections become increasingly difficult or impossible to treat.
The study, which appears in Nature Communications, has found that the use of nanoparticles in combination with other antibiotics, is an effective strategy to improve bacterial killing.
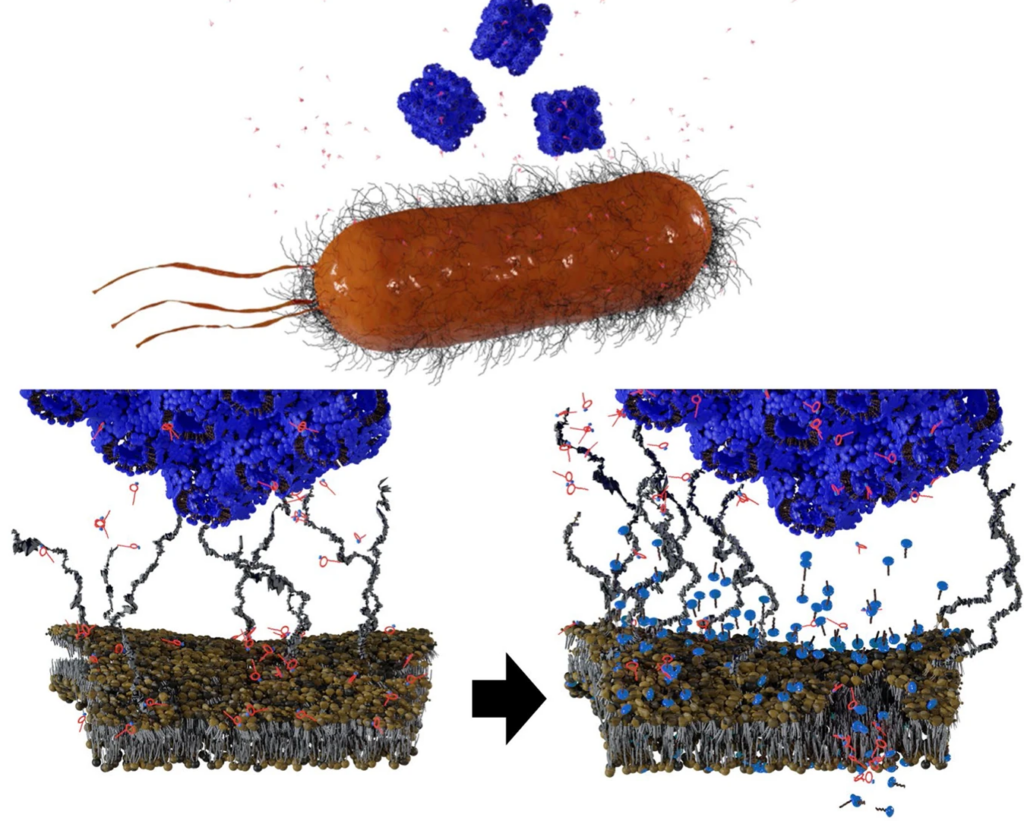
For Gram-negative bacteria, polymyxins have been used as drugs of last resort as they disrupt the bacterial outer membrane (OM), causing it to become more permeable, causing cell contents to leak out and kill the bacteria.
The strategy involves administering polymyxin B (PMB) alongside cube-shaped nanoparticles called cubosomes. The PMB disrupted the OM first, but not enough to kill the cell. When the accompanying cubosome bound to the OM, disrupting it further, successfully killing the cell. Interestingly, loading PMB into the cubosomes as a carrier had little effect; in fact, the cubosome strengthened the OM.
“This is a stunning finding in how we deliver medicine and how the medicine we take impacts us in the future,” said lead researcher Dr Hsin-Hui Shen.
This approach also means that lower dosages of antibiotics could be used. “Instead of looking for new antibiotics to counteract superbugs, we can use the nanotechnology approach to reduce the dose of antibiotic intake, effectively killing multidrug-resistant organisms.”
It has been 30 years since the discovery of the last new antibiotic, and in coming years, the growing crisis of antibiotics resistance will result in increased mortality from basic infections because they have developed antimicrobial resistance.
Without effective antimicrobials, the WHO warns that the success of modern medicine in treating infections, including during major surgery and cancer chemotherapy, would be at increased risk.
While nanoparticles had been used for a long time before as antimicrobial carriers, “but the use of nanoparticles in polytherapy treatments with antibiotics in order to overcome antimicrobial resistance has been overlooked,” explained Dr Shen. “The use of nanoparticles-antibiotics combination therapy could reduce the dose intake in the human body and overcome the multidrug resistance.”
Research will now progress to the testing phase.
Source: Monash University