New Enzymatic Cocktail can Kill Tuberculosis-causing Mycobacteria
With resistance to chemical antibiotics on the rise, the world needs entirely new forms of antibiotics. A new study published in Microbiology Spectrum, a journal of the American Society for Microbiology, shows that an enzymatic cocktail can kill a variety of mycobacterial species of bacteria, including those that cause tuberculosis. The research was carried out by scientists at Colorado State University and Endolytix Technologies.
“We have a mycobacterial drug that works for Nontuberculous Mycobacteria and M. tuberculosis that is biological, not phage therapy, and not small molecule antibiotics,” said Jason Holder, Ph.D., a study coauthor and Founder and Chief Science Officer at Endolytix Technology.
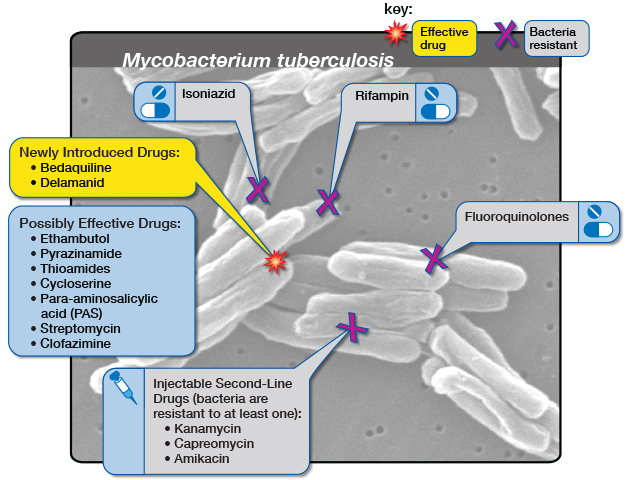
“Mycobacterial infections are particularly hard to treat due to poor efficacy with standard of care drugs that are used in multidrug regimens resulting in significant toxicities and treatments lasting 6 months to years. This is often followed up by reemergence of the bacterial infection after a year of testing negative.”
In the new proof of principle study, the researchers took a biological approach instead of a chemical one to develop a cocktail of enzymes that attack the cell envelope of mycobacteria.
The cocktail of enzymes contains highly specific biochemical catalysts that target and degrade the mycobacteria cell envelope that is essential for mycobacterial viability.
To increase efficacy, the researchers delivered the enzymatic drug inside of host macrophages where mycobacteria grow. In laboratory experiments, the drug was effective against M. tuberculosis and Nontuberculous Mycobacteria (NTMs), both lethal pulmonary lung diseases (PD). TB kills roughly 1.5 million people per year.
“We characterised the mechanism of bactericide as through shredding of the bacterial cells into fragments,” Holder said.
“We’ve shown we can design and develop biological antibiotics and deliver them to the sites of infection through liposomal encapsulation. By combining drug delivery science with enzymes that lyse bacteria, we hope to open up treatment options in diseases such as NTM pulmonary disease, tuberculosis pulmonary disease and others.”
According to study coauthor Richard Slayden, PhD, a professor in the Department of Microbiology, Immunology and Pathology at Colorado State University, the new therapy complements current standard-of-care drugs and does not have many of the drug-drug interactions that are problematic with many anti-mycobacterial drugs in use. “Endolytix enzymes work powerfully with standard-of-care antibiotics to kill bacteria with lower drug concentrations,” Holder said. “This has the potential to reduce the significant toxicities associated with multi-drug regimens that are the standard for mycobacterial infections and hopefully lead to more rapid cures.”